Jim Bednar writes:
"We are pleased to announce the availability of a comprehensive new example of modelling topographic maps in the visual cortex, suitable as a ready-to-run starting point for future research. This example consists of:
1. A new J. Neuroscience paper (Stevens et al. 2013a) describing the GCAL model and showing that is is stable, robust, and adaptive, developing orientation maps like those observed in ferret V1.
2. A new open-source Python software package, Lancet, for launching simulations and collating the results into publishable figures.
3. A new Frontiers in Neuroinformatics paper (Stevens et al. 2013b) describing a lightweight and practical workflow for doing reproducible research using Lancet and IPython.
4. An IPython notebook showing the precise steps necessary to reproduce the 842 simulation runs required to reproduce the complete set of figures and text of Stevens et al. (2013a), using the Topographica simulator.
5. A family of Python packages that were once part of Topographica but are now usable by a broader audience. These packages include 'param' for specifying parameters declaratively, 'imagen' for defining 0D, 1D, and 2D distributions (such as visual stimuli), and 'featuremapper' for analyzing the activity of neural populations (e.g. to estimate receptive fields, feature maps, or tuning curves).
The resulting recipe for building mechanistic models of cortical map development should be an excellent way for new researchers to start doing work in this area.
Jean-Luc R. Stevens, Judith S. Law, Jan Antolik, Philipp Rudiger, Chris Ball, and James A. Bednar
Computational Systems Neuroscience Group
The University of Edinburgh
_______________________________________________________________________________
Jean-Luc R. Stevens, Judith S. Law, Jan Antolik, and James A. Bednar. Mechanisms for stable, robust, and adaptive development of orientation maps in the primary visual cortex. Journal of Neuroscience, 33:15747-15766, 2013. http://dx.doi.org/10.1523/JNEUROSCI.1037-13.2013
shows how the GCAL model was designed to replace previous models of V1development that were unstable and not robust. The model in this paper accurately reproduces the process of orientation map development in ferrets, as illustrated in this animation comparing GCAL, a simpler model, and chronic optical imaging data from ferrets.
run jobs, and collate the output from an external simulator or analysis tool. The approach is fully general, to allow the researcher to switch between different software tools and platforms as necessary.
Lancet is designed to integrate well into an exploratory workflow within the Notebook environment offered by the IPython project. In an IPython notebook, you can generate data, carry out analyses, and plot the results interactively, with a complete record of all the code used.
Together with Lancet, it becomes practical to automate every step needed to generate a publication within IPython Notebook, concisely and reproducibly. This new paper describes the reproducible workflow and shows how to use it in your own projects.
needed to reproduce all the published figures in the paper. Static copies of these notebooks, along with instructions for downloading runnable versions, can be viewed here:
http://topographica.org/_static/gcal.html
http://topographica.org/_static/stevens_jn13.html
param: The parameters offered by param allow scientific Python programs to be written declaratively, with type and range checking, optional documentation strings, dynamically generated values, default values and many other features.
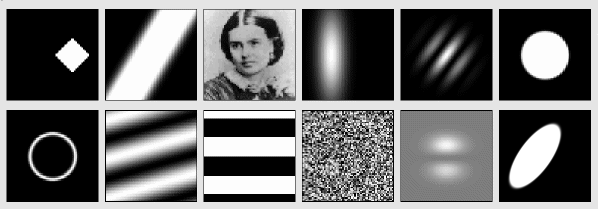
imagen: Imagen offers a set of 0D,1D and 2D pattern distributions. These patterns may be procedurally generated or loaded from files. They can be used to generate simple scalar values, such as values drawn from a specific random distribution, or for generating complex, resolution-independent composite image pattern distributions typically used as visual stimuli (e.g. Gabor and Gaussian patches or masked sinusoidal gratings).
featuremapper: Featuremapper allows the response properties of a neural population to be measured from any simulator or experimental setup that can give estimates of the neural activity values in response to an input pattern. Featuremapper may be used to measure preference and selectivity maps for various stimulus features (e.g orientation and direction of visual stimuli, or frequency for auditory stimuli), to compute tuning curves for these features, or to measure receptive fields, regardless of the underlying implementation of the model or experimental setup."
"We are pleased to announce the availability of a comprehensive new example of modelling topographic maps in the visual cortex, suitable as a ready-to-run starting point for future research. This example consists of:
1. A new J. Neuroscience paper (Stevens et al. 2013a) describing the GCAL model and showing that is is stable, robust, and adaptive, developing orientation maps like those observed in ferret V1.
2. A new open-source Python software package, Lancet, for launching simulations and collating the results into publishable figures.
3. A new Frontiers in Neuroinformatics paper (Stevens et al. 2013b) describing a lightweight and practical workflow for doing reproducible research using Lancet and IPython.
4. An IPython notebook showing the precise steps necessary to reproduce the 842 simulation runs required to reproduce the complete set of figures and text of Stevens et al. (2013a), using the Topographica simulator.
5. A family of Python packages that were once part of Topographica but are now usable by a broader audience. These packages include 'param' for specifying parameters declaratively, 'imagen' for defining 0D, 1D, and 2D distributions (such as visual stimuli), and 'featuremapper' for analyzing the activity of neural populations (e.g. to estimate receptive fields, feature maps, or tuning curves).
The resulting recipe for building mechanistic models of cortical map development should be an excellent way for new researchers to start doing work in this area.
Jean-Luc R. Stevens, Judith S. Law, Jan Antolik, Philipp Rudiger, Chris Ball, and James A. Bednar
Computational Systems Neuroscience Group
The University of Edinburgh
_______________________________________________________________________________
1. STEVENS et. al. 2013a: GCAL model
Our recent paper:Jean-Luc R. Stevens, Judith S. Law, Jan Antolik, and James A. Bednar. Mechanisms for stable, robust, and adaptive development of orientation maps in the primary visual cortex. Journal of Neuroscience, 33:15747-15766, 2013. http://dx.doi.org/10.1523/JNEUROSCI.1037-13.2013
shows how the GCAL model was designed to replace previous models of V1development that were unstable and not robust. The model in this paper accurately reproduces the process of orientation map development in ferrets, as illustrated in this animation comparing GCAL, a simpler model, and chronic optical imaging data from ferrets.
2. LANCET
Lancet is a lightweight Python package that offers a set of flexible components to allow researchers to declare their intentions succinctly and reproducibly. Lancet makes it easy to specify a parameter space,run jobs, and collate the output from an external simulator or analysis tool. The approach is fully general, to allow the researcher to switch between different software tools and platforms as necessary.
3. STEVENS et. al. 2013b: LANCET/IPYTHON workflow
Jean-Luc R. Stevens, Marco I. Elver, and James A. Bednar. An Automated and Reproducible Workflow for Running and Analyzing Neural Simulations Using Lancet and IPython Notebook. Frontiers in Neuroinformatics, in press, 2013. http://www.frontiersin.org/Journal/10.3389/fninf.2013.00044/abstractLancet is designed to integrate well into an exploratory workflow within the Notebook environment offered by the IPython project. In an IPython notebook, you can generate data, carry out analyses, and plot the results interactively, with a complete record of all the code used.
Together with Lancet, it becomes practical to automate every step needed to generate a publication within IPython Notebook, concisely and reproducibly. This new paper describes the reproducible workflow and shows how to use it in your own projects.
4. NOTEBOOKS for Stevens et al. 2013a
As an extended example of how to use Lancet with IPython to do reproducible research, the complete recipe for reproducing Stevens et al. 2013a is available in models/stevens.jn13 of Topographica's GitHub repository. The first of two notebooks defines the model, alternating between code specification, a textual description of the key model properties with figures, and interactive visualization of the model's initial weights and training stimuli. The second notebook can be run to quickly generate the last three published figures (at half resolution) but can also launch all 842, high-quality simulationsneeded to reproduce all the published figures in the paper. Static copies of these notebooks, along with instructions for downloading runnable versions, can be viewed here:
http://topographica.org/_static/gcal.html
http://topographica.org/_static/stevens_jn13.html
5. PARAM, IMAGEN, and FEATUREMAPPER
The Topographica simulator has been refactored into several fully independent Python projects available on GitHub (http://ioam.github.io). These projects are intended to be useful to a wide audience of both computational and experimental neuroscientists:param: The parameters offered by param allow scientific Python programs to be written declaratively, with type and range checking, optional documentation strings, dynamically generated values, default values and many other features.
imagen: Imagen offers a set of 0D,1D and 2D pattern distributions. These patterns may be procedurally generated or loaded from files. They can be used to generate simple scalar values, such as values drawn from a specific random distribution, or for generating complex, resolution-independent composite image pattern distributions typically used as visual stimuli (e.g. Gabor and Gaussian patches or masked sinusoidal gratings).
featuremapper: Featuremapper allows the response properties of a neural population to be measured from any simulator or experimental setup that can give estimates of the neural activity values in response to an input pattern. Featuremapper may be used to measure preference and selectivity maps for various stimulus features (e.g orientation and direction of visual stimuli, or frequency for auditory stimuli), to compute tuning curves for these features, or to measure receptive fields, regardless of the underlying implementation of the model or experimental setup."